Observing and Analyzing How Ribosomes Respond to Perturbations
Using cryo-electron microscopy (cryo-EM), I focus on observing the structure of ribosomes to reveal their dynamic features, responding to perturbations such as antibiotic binding.
The ribosome is the major target of antibiotics. Our DARC method visualizes how antibiotics act on ribosomes in a molecular crowding environment.

TAKESHI YOKOYAMA Ph.D.
Assistant Professor,
Department of Life Sciences,
The Advanced Research Center for Innovations in Next-Generation Medicine (INGEM),
Tohoku University
Selected Publications
Yuka Isozaki, Takumi Makikawa, Kosuke Kimura, Daiki Nishihara, Maho Fujino, Yoshikazu Tanaka, Chigusa Hayashi, Yoshimasa Ishizaki, Masayuki Igarashi*, Takeshi Yokoyama*, Kazunobu Toshima*, Daisuke Takahashi*.
Creation of a macrolide antibiotic against non-tuberculous Mycobacterium using late-stage boron-mediated aglycon delivery
Science Advances 11(10) adt2352
doi: 10.1126/sciadv.adt2352
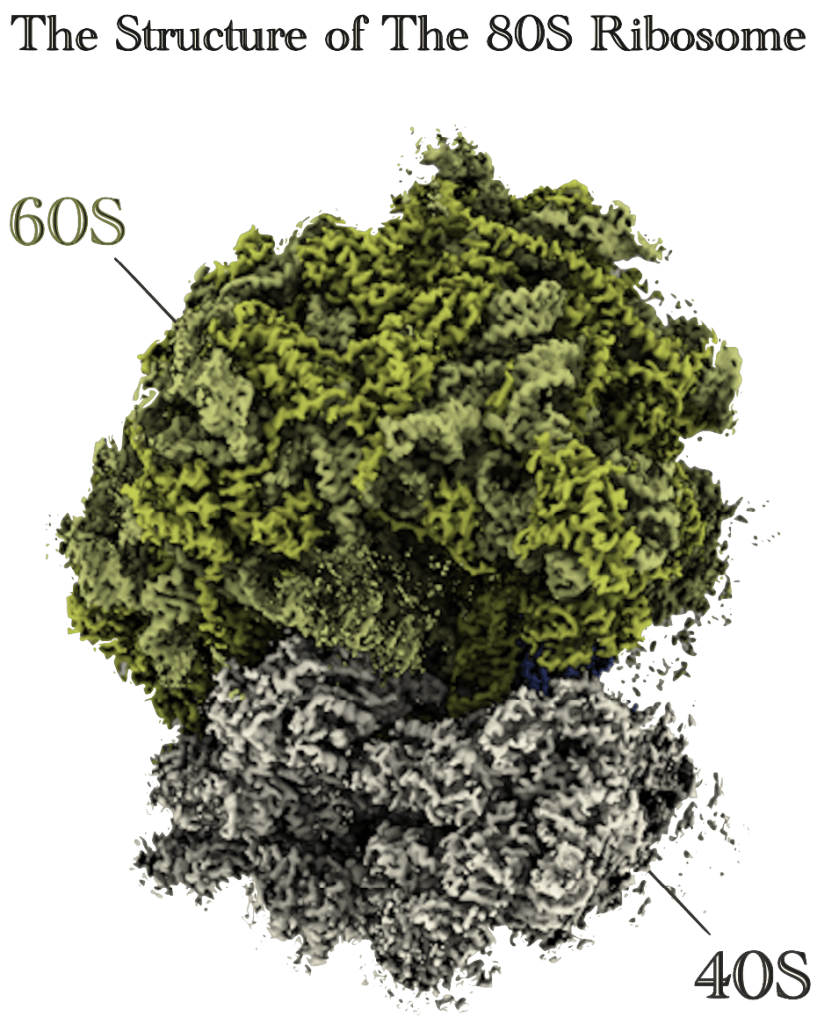
Mayuki Tanaka#, Takeshi Yokoyama#, Hironori Saito#, Madoka Nishimoto, Kengo Tsuda, Naoyuki Sotta, Hideki Shigematsu, Mikako Shirouzu, Shintaro Iwasaki*, Takuhiro Ito*, and Toru Fujiwara*.
Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1 at the A site.
Nature Chemical Biology 20(5): 605-614 (2024)
doi: 10.1038/s41589-023-01513-0
Junta Tomono#, Kosuke Asano#, Takuma Chiashi, Masato Suzuki, Masayuki Igarashi, Yoshiaki Takahashi, Yoshikazu Tanaka*, and Takeshi Yokoyama*.
Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
The Journal of Biochemistry
175(6): 587-598 (2024)
doi: 10.1093/jb/mvae002
Naho Akiyama, Kensuke Ishiguro, Takeshi Yokoyama, Kenjyo Miyauchi, Asuteka Nagao, Mikako Shirouzu, and Tsutomu Suzuki*.
Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine.
Nature Structural & Molecular Biology
31(5): 817-825 (2024)
doi: 10.1038/s41594-024-01238-1
Xuewei Zhao, Ding Ma, Kensuke Ishiguro, Hironori Saito, Shinichiro Akichika, Ikuya Matsuzawa, Mari Mito, Toru Irie, Kota Ishibashi, Kimi Wakabayashi, Yuriko Sakaguchi, Takeshi Yokoyama, Yuichiro Mishima, Mikako Shirouzu, Shintaro Iwasaki, Takeo Suzuki,* and Tsutomu Suzuki*.
Glycosylation of queuosine in tRNAs contributes to optimal translation and post-embryonic growth in vertebrates.
Cell
186(25) 5517-5535.e24 (2023) doi: 10.1016/j.cell.2023.10.026
Chisa Shiraishi, Akinobu Matsumoto, Kazuya Ichihara, Taishi Yamamoto, Takeshi Yokoyama, Taisuke Mizoo, Atsushi Hatano, Masaki Matsumoto, Yoshikazu Tanaka, Eriko Matsuura-Suzuki, Shintaro Iwasaki, Shouji Matsushima, Hiroyuki Tsutsui, Keiichi I.
Nakayama*.RPL3L-containing ribosomes determine translation elongation dynamics required for cardiac function.
Nature Communications
14(1):2131 (2023)
doi: 10.1038/s41467-023-37838-6
Yuko Murayama, Haruhiko Ehara, Mari Aoki, Mie Goto, Takeshi Yokoyama, Shun-ichi Sekine*.
Structural basis of the transcription termination factor Rho engagement with transcribing RNA polymerase.
Science Advances
9(6) eade7093 (2023)
doi: 10.1126/sciadv.ade7093
Hiromi Watari#, Hiromu Kageyama#, Nami Masubuchi, Hiroya Nakajima, Kako Onodera, Pamela J. Focia, Takumi Oshiro, Takashi Matsui, Yoshio Kodera, Tomohisa Ogawa, Takeshi Yokoyama, Makoto Hirayama, Kanji Hori, Douglas M. Freymann, Misa Imai, Norio Komatsu, Marito Araki*, Yoshikazu Tanaka*, Ryuichi Sakai*.
A marine sponge-derived lectin reveals hidden pathway for thrombopoietin receptor activation.
Nature Communications
13(1) (2022)
doi: 10.1038/s41467-022-34921-2
Yoshiko Nakagawa, Howard C.-H. Shen, Yusuke Komi, Shinju Sugiyama, Takaaki Kurinomaru, Yuri Tomabechi, Elena Krayukhina, Kenji Okamoto, Takeshi Yokoyama, Mikako Shirouzu, Susumu Uchiyama, Megumi Inaba, Tatsuya Niwa, Yasushi Sako, Hideki Taguchi, Motomasa Tanaka*.
Amyloid conformation-dependent disaggregation in a reconstituted yeast prion system.
Nature Chemical Biology
Mar;18(3):321-331. (2022)
doi: 10.1038/s41589-021-00951-y
Mutsuko Kukimoto-Niino, Kazushige Katsura, Rahul Kaushik, Haruhiko Ehara, Takeshi Yokoyama, Tomomi Uchikubo-Kamo, Reiko Nakagawa, Chiemi Mishima-Tsumagari, Mayumi Yonemochi, Mariko Ikeda, Kazuharu Hanada, Kam Y. J. Zhang, Mikako Shirouzu*.
Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex.
Science Advances
7(30) eabg3147 (2021)
doi: 10.1126/sciadv.abg3147
Takeshi Yokoyama#, Kodai Machida#, Wakana Iwasaki#, Tomoaki Shigeta, Madoka Nishimoto, Mari Takahashi, Ayako Sakamoto, Mayumi Yonemochi, Yoshie Harada, Hideki Shigematsu, Mikako Shirouzu, Hisashi Tadakuma*, Hiroaki Imataka*, Takuhiro Ito*.
HCV IRES Captures an Actively Translating 80S Ribosome
Molecular Cell
74(6) 1205-1214.e8 (2019)
doi: 10.1016/j.molcel.2019.04.022
Yongchan Lee, Pattama Wiriyasermkul, Chunhuan Jin, Lili Quan, Ryuichi Ohgaki, Suguru Okuda, Tsukasa Kusakizako, Tomohiro Nishizawa, Kazumasa Oda, Ryuichiro Ishitani, Takeshi Yokoyama, Takanori Nakane, Mikako Shirouzu, Hitoshi Endou, Shushi Nagamori, Yoshikatsu Kanai, Osamu Nureki*.
Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc.
Nature Structural & Molecular Biology
26(6) 510-517 (2019)
doi: 10.1038/s41594-019-0237-7
Kazuhiro Kashiwagi, Takeshi Yokoyama, Madoka Nishimoto, Mari Takahashi, Ayako Sakamoto, Mayumi Yonemochi, Mikako Shirouzu, Takuhiro Ito*.
Structural basis for eIF2B inhibition in integrated stress response.
Science
364(6439) 495-499 (2019)
doi: 10.1126/science.aaw4104
Go Kasuya#, Takanori Nakane#, Takeshi Yokoyama#, Yanyan Jia, Masato Inoue, Kengo Watanabe, Ryoki Nakamura, Tomohiro Nishizawa, Tsukasa Kusakizako, Akihisa Tsutsumi, Haruaki Yanagisawa, Naoshi Dohmae, Motoyuki Hattori, Hidenori Ichijo, Zhiqiang Yan, Masahide Kikkawa, Mikako Shirouzu, Ryuichiro Ishitani*, Osamu Nureki*
Cryo-EM structures of the human volume-regulated anion channel LRRC8
Nature Structural & Molecular Biology
25(9) 797-804 (2018)
doi: 10.1038/s41594-018-0109-6
Haruhiko Ehara, Takeshi Yokoyama, Hideki Shigematsu, Shigeyuki Yokoyama, Mikako Shirouzu, Shun-ichi Sekine*.
Structure of the complete elongation complex of RNA polymerase II with basal factors.
Science
357(6354) 921-924 (2017)
doi: 10.1126/science.aan8552
Takeshi Yokoyama, Tsutomu Suzuki*.
Ribosomal RNAs are tolerant toward genetic insertions: evolutionary origin of the expansion segments.
Nucleic Acids Research
36(11) 3539-3551 (2008)
doi: 10.1093/nar/gkn224
Research map
https://researchmap.jp/yokotake
ORCID
DARC Method Visualizes Antibiotic Action on Ribosome
Recently, we developed a method to directly observe the dynamic structures of ribosomes in molecular crowding environments using cryo-electron microscopy (cryo-EM).
By utilizing this new technique, we aim to visualize the function of antibiotics, with the goal of developing new antibiotics to counter the growing issue of antimicrobial resistance (AMR).

The DARC method: Direct Visualization of Antibiotic binding on Ribosomes in the Cell-free translation system
(Tomono, Asano, et al, J Biochem, 2024)
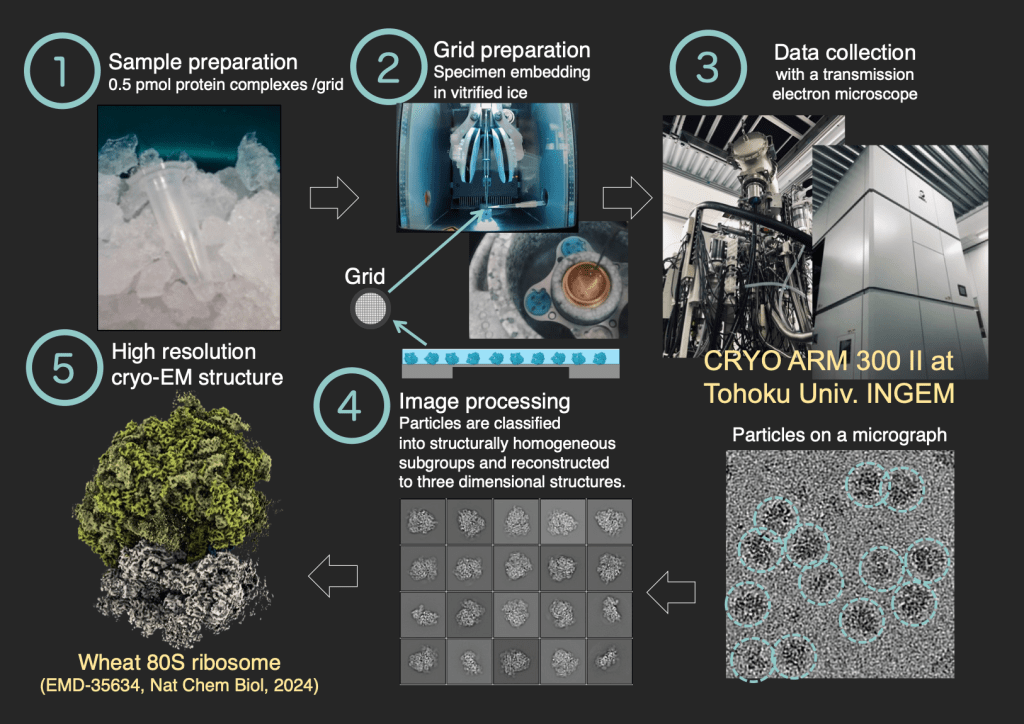
Cryo-Electron Microscopy at Tohoku Univ.
We are using the CRYO ARM 300 II (JEOL), introduced at Tohoku University, to facilitate numerous collaborative research projects. Our structural analysis work extends beyond ribosomes, aiming to achieve high-resolution structural analysis of diverse biological specimens.